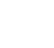Predictive Design of Biologics
Rapid Computational Design & Experimental Validation of Antibodies
LLNL biologists, computer scientists and engineers are developing a machine-learning enabled computational/experimental framework that enables unprecedented acceleration of the drug development process.
Our platform integrates world-class high-performance computing and simulation, machine learning, structural bioinformatics, and multi-objective optimization to perform in silico antibody and antigen design and optimization. These designs are then experimentally validated at LLNL and at our partner facilities, and results are either pursed as candidate medical countermeasures (MCMs) or provided as critical feedback for computational redesign.
We are partnering with academia, and industry to further develop and demonstrate this capability. When fully implemented our system will enable both a proactive and rapid reactive approach for designing MCMs
If you are interested in joining our team or have questions, please contact us at CPDB [at] llnl.gov (CPDB[at]llnl[dot]gov)
Research Highlights
Computationally restoring the potency of a clinical antibody against SARS-CoV-2 Omicron subvariants
With the emergence of the SARS-CoV-2 Omicron variant in late 2021, many clinically used antibody drug products lost potency, including Evusheld and its constituent, cilgavimab. Rapidly modifying such high-value antibodies with a known clinical profile to restore efficacy against emerging variants is a compelling mitigation strategy. We sought to redesign COV2-2130 to rescue in vivo efficacy against Omicron BA.1 and BA.1.1 strains while maintaining efficacy against the contemporaneously dominant Delta variant. Here we show that our computationally redesigned antibody, 2130-1-0114-112, achieves this objective, simultaneously increases neutralization potency against Delta and many variants of concern that subsequently emerged, and provides protection in vivo against the strains tested.

AbBERT: Learning Antibody Humanness Via Masked Language Modeling
One component of our computational pipeline is a model to understand the degree of similarity of novel, engineered antibody sequences to typical human antibody sequences. Accurate understanding of the degree of humanness can help reduce or manage the risk of failure modes such as immunogenicity or poor manufacturability. AbBERT was developed to fill this need as a transformer-based language model derived from ProtBert, which was trained on up to 20 million unpaired sequences from the Observed Antibody Space database.
We validated AbBERT using a novel “multi-mask'' scoring procedure to demonstrate high accuracy in predicting antibody complementary determining regions—including the challenging hypervariable H3 region. We then demonstrated several uses of AbBERT on downstream antibody design and prediction tasks. AbBERT enhances in silico antibody optimization via deep reinforcement learning by utilizing its learned embeddings as additional observations during optimization.
Other Research Manuscripts
- SARS-COV-2 Omicron variant predicted to exhibit higher affinity to ACE-2 receptor and lower affinity to a large range of neutralizing antibodies, using a rapid computational platform
- Language model-accelerated deep symbolic optimization
- Learning Regions of Interest for Bayesian Optimization with Adaptive Level-Set Estimation
- Large-scale application of free energy perturbation calculations for antibody design
- Learning Regions of Interest for Bayesian Optimization with Adaptive Level-Set Estimation
Join Our Team

We have career opportunities to develop a next generation computational antibody and antigen design platform driven by simulation and machine learning models trained on large experimental datasets.
Lawrence Livermore National Laboratory has been ranked among the top 10 employers in the San Francisco Bay Area by Forbes Magazine, won multiple Glassdoor Best Places to Work awards, and is among the top 12 in government services nationwide. The Laboratory offers employees outstanding careers with a first-class benefits package in a beautiful location. See our Careers page for more information about our culture, the benefits and perks, our history, and job opportunities.
Our Leadership

Dan Faissol, PhD
Principal Investigator

Shankar Sundaram, PhD
Program Executive

Tom Bates, PhD
Program Deputy

John Hernandez
Business Project Manager

Kate Arrildt, PhD
Virology, Vaccinology

Thomas Bunt
Rapid Response Laboratory Lead

Barry Chen, PhD
Language Modeling Lead

Simone Conti, PhD
Molecular Dynamics Simulations

Jie Deng
Software and Data Architect

Tom Desautels, PhD
Multi-fidelity Simulation and Machine Learning for Efficacy Prediction

John Goforth
Software Development Lead

Mikel Landajuela, PhD
Optimization Lead

Ed Lau, PhD
Molecular Dynamics Simulation

Ed Saada, PhD
Pathogen Interactions and Medical Countermeasures

Ana Paula Sales, PhD
Developability Prediction Lead

Brent Segelke, PhD
Structural Biologist, Protein Biochemist

Adam Zemla, PhD
Protein Structure Bioinformatics

Fangqiang Zhu, PhD
Molecular Dynamics Simulation, Structural Antibody Modeling
The Team

Aram Avila-Herrera
Bioinformatics Software Developer

Drew Bennett, PhD
Molecular Dynamics Simulation, Multi-scale Modeling

Jose Cadena Pico, PhD
Machine Learning

Ippolito Imani Caradonna
Automation Expert

Rudrasis Chakraborty
Researcher

Matt Coleman, PhD
Vaccinologist, Biochemical Engineer

Zachary Cosenza
Bayesian Modeling, Multi-task Learning

Aneesha Devulapally
ChemBio Data Scientist

Jeff Drocco, PhD
Bioinformatics

Andre Goncalves, PhD
Machine Learning, Multi-task Learning, Multi-objective Optimization

Emilia Grzesiak
Biostatics and Data Science, Software Development

Rebecca Haluska
Software Development and Database Engineering

Conor Hayes, PhD
Reinforcement Learning, Machine Learning

Matthew Jemielita
Assay Dev Lead

Piyush Karande, PhD
Deep Learning

Tek Hyung Lee, PhD
Synthetic and Systems Biologist and Microbial Engineer

Leno De Silva, PhD
Deep Reinforcement Learning

Tavish McDonald
Machine Learning, Language Modeling

Yamen Mubarka
Deep Learning

Nate Mundhenk, PhD
Deep learning, Convolutional Neural Networks

Rami Musharrafieh
Vaccines Lead

Christine Ocampo
Operation Administrator

Kim Phan
Administrative Specialist

Hiranmayi Ranganathan, PhD
Deep Active Learning, Machine Learning

Tiffany Reck
Administrative Specialist

Dante Ricci, PhD
Molecular Genetics, Systems Biology

Natalie Rowland
Administrative Specialist

Bonnee Rubinfeld
Cellular, Molecular, Micro-biologist/Biochemist

Mary Silva
Bayesian Statistics and Data Science

Cheryl Strout
Biochemical and Analytical Scientist

Gareth Trubl, PhD
Viral and Microbial Ecologist

Camilo Valdes, PhD
Bioinformatics

Denis Vashchenko
Deep Learning Language Modeling, Software Development
Yaqing Wang, PhD
Structural Biologist, Molecular Biophysicist

Dina Weilhammer, PhD
Immunologist, Virologist

Tracy Weisenberger
Cellular, Molecular, and Viral Biologist

Boya Zhang, PhD
Machine learning, surrogate-based modeling

Interested in joining the cutting edge of computational design and bioengineering?
CPDB [at] llnl.gov (Contact us )